| Contents |
|
- General
- Selecting a species and a genome dataset
- Entering the protein query
- Setting Scipio's search options
- Viewing Scipio's results
- Scaling in gene pictures
| General | Back to top |
To obtain the gene structure corresponding to a certain protein query, first select a species and a genome dataset, then enter your protein query, optionally adjust Scipio's settings, and finally start Scipio.
| Selecting a species and a genome dataset | Back to top |
Species are selected using an autocompletion form. Either scientific names, common names, or taxa may be entered. Note, that fungi often have different names for their teleomorph and anamorph. Both names can be searched for, but only the anamorph name will be listed at the moment. Strains will be listed separately, Typing a taxon-name is very useful, if the user wants to perform a cross-species search and is looking for closely related species. In the example, mamm has been typed, the first letters of Mammalia, and the autocompletion lists the first 10 species out of 50 mammalian species for which genome data is available.
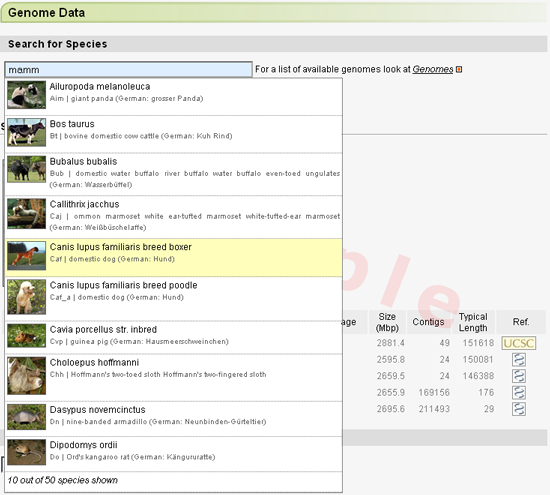
As soon as a species has been selected, the available genome datasets will be listed. Different version numbers refer to the different version of the genome assemblies. Note, that the larger the file the longer the search will take. For many species, contig and supercontig data is available. For some species, the supercontigs have already been ordered into chromosomes. Uchromosomes contain the contigs that could not be placed onto chromosomes yet.
| Entering the protein query | Back to top |
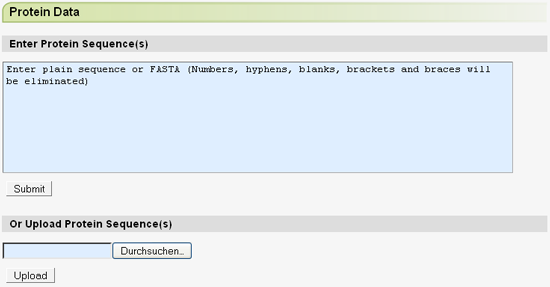
When a genome dataset has been chosen, the protein query can be entered and Scipio and BLAT options can be set. The protein query can be entered in either plain text or FASTA format.
| Setting Scipio's search options | Back to top |
Now, Scipio can be started either using the default settings (maximum mismatch will be 7) that are most suitable for in-species searches, or several options might be set to allow for e.g. more mismatches or to use a smaller BLAT-tilesize to also get very small exons. To set those options, open the "Expert Options" menu:
Scipio Options
Min. Score
Minimal score (between 0 and 1) of a hit for a query, in order to appear in the output. The formula for the score is: matches minus mismatches, devided by query length. If a hit is composed of multiple partial hits, the minimal score applies to the best-scoring partial hit. (default: 0.3) E.g. a Min. Score of 0.3 means, that at least 30 % of the protein query must be found on one contig.
Min. Identity
The minimal identity between query and target sequence in every single BLAT hit to be processed by Scipio, in percent (default is 90).
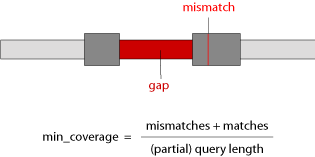
Min. Coverage
The Min. Coverage is the minimal portion of the respective query sequence that must be found in every single BLAT hit to be processed by Scipio, in percent (default is 60). In the case of partial hits, we consider only the part between the first and the last residue of the query, that is aligned to the target sequence, for calculating the coverage. Gaps at the query start or end, or between partial hits have no influence on the coverage. This parameter has been introduced to exclude hits from the assembly that by chance show homology, that is recognized by Blat, in very short stretches although most of the query sequence could not be found.
 |
Max. Mismatch
The maximum number of mismatches allowed on a contig in order to be included in the results. If the result consists of the assembly of more than one contig, the allowed maximum number of mismatches is the given value times the number of contigs. The Value "∞" allows an unlimited number of mismatches. If another translation table than (1), The Standard Code, is selected, then it is not possible to change this value.
Region Size
The length of the up- and downstream regions that will be retrieved.
Multiple Results
Sometimes, there are gene duplicates in the genome resulting from whole genome or single gene duplications. Setting the Multiple Results option to Yes, all hits for the same query with scores exceeding the minimal score will be shown. Multiple hits are named <queryname>_(1), <queryname>_(2), etc.
Single Target Hits Only
By turning this option to On, Scipio will not assemble hits on multiple targets/contigs. It is recommended to use this option for chromosome target files.
Rationale:
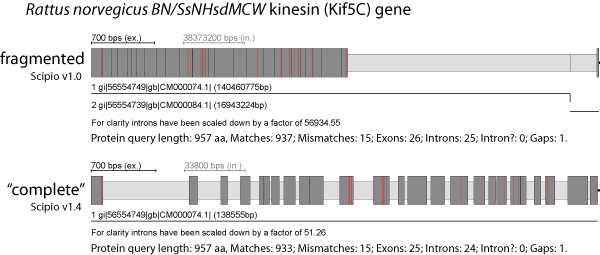
Scipio is able to reconstruct genes that are spread on several contigs or supercontigs of highly fragmented genomes. As we have shown, this feature is one of the most important strengths of Scipio that other programs do not offer. However, this feature is not needed in chromosomal assemblies, and might lead, especially in the case of cross-species searches, to composed hits that stretch across multiple chromosomes, one of them false positive (Figure). Hence, it can be switched off with the parameter --single_target_hits.
The figure shows an example of the search for the rat homolog (target sequence) of the human Kif5C kinesin motor protein. The C-terminal about 25 amino acids of the rat Kif5C homolog are missing in the respective chromosome assembly. Using Scipio v1.0 a very short identical stretch of four amino acids, found on a different chromosome, has artificially been added to the 3′-end of the gene generating an "intron" of millions of base pairs (Note the scale of the introns!). The new parameters --single_target_hits and --max_assemble_size (see below) now prevent this mis-assembly.
 |
Translation Table
Due to the different codon translations of some organisms (e.g. Candida species, Tetrahymena thermophila, etc.) the use of the standard translation table leads to many mismatches.
For more information about translation tables see:
![]() The Genetic Codes
The Genetic Codes
Scipio Expert Options
Max. Assemble Size
Max. Assemble Size is the maximum size of intron parts at target boundaries. If an intron would have to be created between two partial hits across two contigs, that exceeds the given size (default: 75000 nucleotides), the two hits cannot appear together as parts of one composed hit. The contig with the lower score will be rejected (unless the score exceeds --min_score, and --multiple_results is enabled). This parameter is very similar to the --single_target_hits option. However, for highly fragmented genomes it is still reasonable to allow gene reconstructions across several contigs. But also in this case one would want to exclude the assembly of hits that would introduce extremely long introns between exons on different contigs. To accomplish for those cases we have introduced the --max_assemble_size parameter.
Max. Move Exon
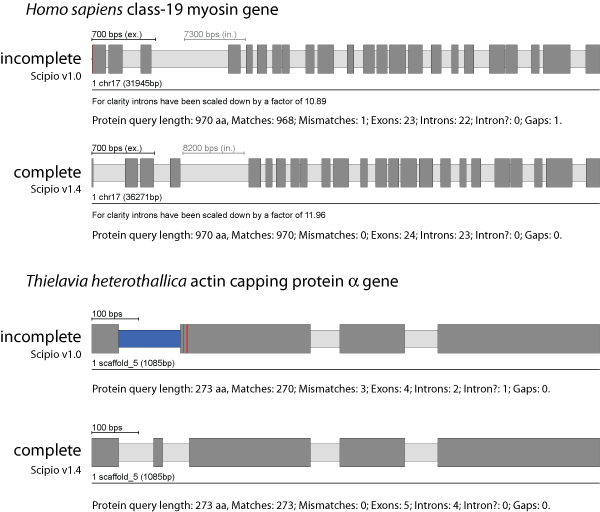
This option determines how much Scipio shifts the BLAT prediction at intron borders to reconstruct correct introns (default: 2 amino acids). BLAT chooses between possible intron positions by minimizing mismatches. In rare cases, mostly cross-species alignments, there are several intron candidates causing an equal number of mismatches so that the correct exon borders can only be recognized by matching the splice site consensus. In even rarer cases, the true intron location is more than two codons away from the BLAT prediction which is when you need this option.
In the case of the human class-19 myosin gene, Blat and the old Scipio version were not able to reconstruct the 5′-end of the gene correctly, because the intron in front of the second exon of the gene ends with the translated sequence LFQ that is very homologous to the real sequence LQQ. Blat added these residues to exon 2 albeit introducing a mismatch. With the parameter --max_move_exon set to 3, Scipio is now able to resolve this misalignment and to subsequently identify the correct exon 1. Case B shows the reconstruction of the actin capping protein α from Theileria heterothallica. Here, by chance the intergenic region before exon 3 shows some homology to exon 2 (3 matches and 3 mismatches) and thus the exon 2 sequence was erroneously joined to exon 3. This happened irrespectively of lowering the Blat tilesize or adjusting any of the other Scipio parameters. By setting --max_move_exon to 6, the new version of Scipio is now able to correctly reconstruct the CAPα gene.
You can find more details and data to play with this parameter here: Parameters to account for low homology at intron borders
Gap to Close and Min Intron Length
Gene homologs even from very closely related species are often too divergent to be completely identified by Blat. While the core building block of the proteins and the functional sites are often strongly conserved, low homology is especially found at the surface of the proteins. Thus, loop regions are often sites of amino acid substitutions, insertions of long stretches of residues, and deletions. In addition, since the terminal regions of most proteins are at the surface, they are very divergent. Short stretches of nucleotides whose lengths are multiples of three and whose translations do not result in any in-frame stop codons are most likely to be insertions rather than true introns.
Gap to Close
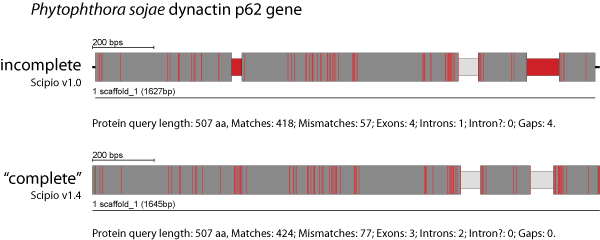
Gap to Close is the maximum size of a gap in a query that Scipio closes by adding mismatches to exon boundaries. By default, up to 6 additional amino acids in the query sequence will be tolerated without introducing a gap (for unmatched query sequence) in the target sequence.
The figure shows an example of divergent homologs of the dynactin p62 gene of Phytophthora ramorum (query sequence) and Phytophthora sojae (target sequence). These two homologs contain a long divergent region with many consecutive mismatches in exon 1 that is not identified by BLAT and introduces a long gap. In addition, the N- and C-termini have divergent sequences and different length. The query sequence is shorter at the termini but the target homolog is correctly reconstructed with Scipio′s --gap_to_close parameter.
You can find more details and data to play with this parameter here: Parameters to account for additional/missing bases in predicted exons
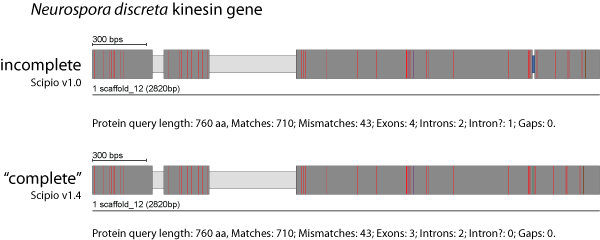
Min. Intron Length
To better model sequence differences that are located within exons we have introduced the new adjustable parameter --min_intron_len. The --min_intron_len parameter has been implemented for those cases that leave additional target sequence after mapping the query sequence. By default, every insertion longer then 21 nucleotides will be treated as intron, while shorter sequences are inserted as additional nucleotides into the surrounding exons, which are then joined to one exon.
The figure shows the results of a search for a kinesin homolog from Neurospora crassa (query sequence) in the closely related organism Neurospora discreta (target sequence). Because of the relativ high homology of the two sequences, BLAT has already retained the additional residues of the query sequence so that they are included in the result of the old Scipio version. However, an intron? was introduced in the region that contained additional nucleotides in the target sequence leading to missing residues in the target translation. With the new parameter --min_intron_len these additional nucleotides are correctly treated as exonic sequence.
You can find more details and data to play with this parameter here: Parameters to account for additional/missing bases in predicted exons
Accepted Splice Sites
GT---AG and GC---AG are by far the most common 3' and 5' splice sites, and thus by default excepted as correct intron borders. In very rare cases, other splice sites like AT---AC, GG---AG, and GA---AG have been observed. To mark these introns as "intron" and not as "intron?" change the Accepted Splice Sites. This parameter does not change the search but the resulting output.
BLAT Options
BLAT Tilesize
Determines the width of the search window used to scan the genome (the size of match that triggers an alignment). Decreasing this value makes it more likely that small exons are found but also slows the search process.
If the genomefile is bigger then 2Gb, the min. tilesize will be limited to 5. If you still want to use a tilesize smaller 5, download scipio and run it on your local machine. For information about the use of the commmand-line tool, consult the documentation. You can also send an email to our group for further help. Use mkollgr@gwdg.de.
BLAT Oneoff
If set to Yes this allows one mismatch in the Blat-tile and still triggers an alignment.
If the genomefile is bigger then 2Gb, the min. tilesize will be limited to 5, and the Blat-oneoff parameter will be disabled if you use 5 as tilesize. If you still want to use oneoff with tilesize 5 or smaller, download Scipio and run it on your local machine. For information about the use of the commmand-line tool, consult the documentation. You can also send an email to our group for further help. Use mkollgr@gwdg.de.
BLAT Min. Score
The Blat score is calculated as matches minus mismatches minus some sort of gap penalty.
BLAT Min. Identity
Sets minimum sequence identity (in percent). By default, --blat_identity is set to 90% of --min_identity (e.g. the default --min_identity is 90, thus the default --blat_identity is 81).
Needlman-Wunsch Options
To identify exons that contain too many mismatches to be identified by Blat, and to correctly annotate very short exons of three or even less amino acids, the Needleman-Wunsch algorithm described above forces an alignment of unmatched query sequence to spare target sequence. The maximal lengths of query and target sequence fragments to be aligned with Needleman-Wunsch are controlled by the parameters --exhaust_align_size and --exhaust_gap_size, respectively. By default, the exhaustive search is restricted to query gaps of at most three times the Blat tilesize, since we expect Blat to successfully discover any longer exons, and to a target subsequence of 500 bps, so that the potentially very long introns in mammalian genomes are only searched after manual interaction. Other parameters affecting the Needleman-Wunsch algorithm, such as the penalties mentioned above, can be adjusted by the commandline verion only, and not via WebScipio.
Exhaust Align Size
The Exhaust Align Size is the maximum sequence length for exhaustive search: because the Needleman-Wunsch algorithm is slow, the optimal alignment will not be computed in DNA regions longer than the given value (default: 500 nucleotides). Instead, Scipio will try here to place additional amino acids at the intron borders (resulting in one big intron), if possible, with few additional mismatches, or otherwise leave some amino acids unmatched (which will appear as "gap").
In the figure, the human dynactin p50 gene contains two very short exons of 3 and 2 amino acids. These two short exons are conserved in all vertebrates. Case B shows the coronin gene from the basidiomycote fungi Puccinia graminis encoding a short 3 amino acid exon. In addition, the codons at the exon/intron junctions of this short exon are split. In most of the other basidiomycotes sequenced so far, this short exon is part of one of the neighbouring exons, or part of a longer exon that includes both neighbouring exons. However, it also exists in the basidiomycote Melampsora laricis-populina. Thus, this short exon is not an artificial creation but a true exon.
You can find more details and data to play with this parameter here: Parameters to identify divergent exons and very short exons
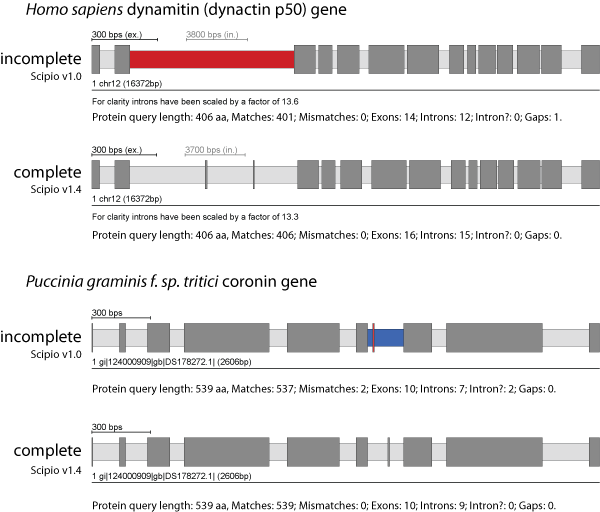
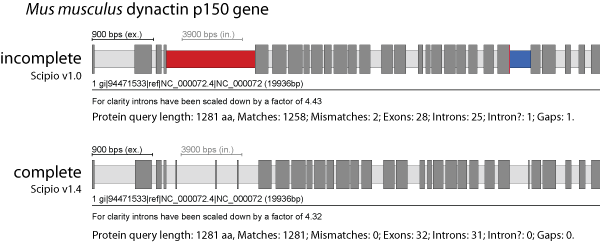
Exhaust Gap Size
The Exhaust Gap Size is the maximum query sequence length (that has not been mapped by Blat) for exhaustive search: because the Needleman-Wunsch algorithm is slow, the optimal alignment will not be computed for query sequences longer than this (default: 3*Blat_tilesize). Instead, Scipio will try here to place additional amino acids at the intron borders (resulting in one big intron), if possible, with few additional mismatches, or otherwise leave some amino acids unmatched (which will appear as "gap").
The figure presents the mouse dynactin p150 gene that contains three short exons of 7, 6, and 7 amino acids at the beginning of the gene. Even with the Blat-tilesize set to 5 those exons are not recognized in the search against the chromosome assembly. Here, the --exhaust_align_size and the --exhaust_gap_size have to be increased to completely reconstruct this part of the gene.
You can find more details and data to play with this parameter here: Parameters to identify divergent exons and very short exons
| Viewing Scipio's results | Back to top |
WebScipio generates a graphical representation of the gene that clearly indicates the length and position of exons and introns and shows where discrepancies are located. It also shows the identifiers of the target sequences. In order not to make small exons vanish when very large intronic stretches are found, the scaling of introns end exons is automatically balanced to make the picture visually meaningful. Tooltips show additional information. For detailed inspection of the hits, WebScipio generates an easy to read alignment of the query and the genome. It is grouped by exons, and mismatches, amino acid insertions and deletions, in-frame stop codons, and frame shifts are highlighted. Different stretches of DNA can be viewed: Up- and downstream DNA, genomic DNA from the first to the last exon including introns, or the coding DNA. The translation of the coding DNA as determined by Scipio can also be viewed. Several types of files can be downloaded: A YAML file which contains all information generated by WebScipio, a GFF file for use with genome software, FASTA files containing all types of DNA sequences (separated or combined), FASTA files containing the protein translation and separated exon translations, the gene structure scheme as SVG in the unscaled or scaled version, and a log file with alignments and detailed evaluation reports.
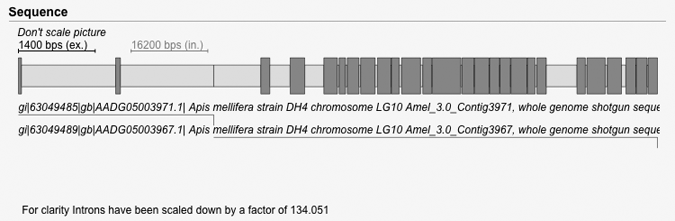
| Scaling in gene pictures | Back to top |
When exons are short compared to introns, the unscaled picture is dominated by introns and it is hard to see the length of the exons (see first picture). To improve the visualization WebScipio scales down the introns and scales up the exons so that the the average length of the introns equal the average length of the exons (see second picture).