| Contents |
|
| General | Back to top |
WebScipio offers an advanced option to map cDNA data (EST or mRNA) on the exon-intron gene structure reconstructed by Scipio. Based on this mapping alternative splice forms are predicted.
| Options | Back to top |
Data
Different cDNA data files can be choosen for different species. An autocompletion field offers the possibility to search for a species. Then an EST or mRNA data file needs to be selected.
Sequence type
In the first step of the cDNA alignment the cDNA data is either aligned to the coding DNA of the gene, the translation of the coding DNA or the whole genomic DNA (including introns). In the second step it is always aligned to the genomic DNA.
BLAT Tile Size
Sets the size of match that triggers an alignment. Usually between 8 and 12. Default is 11 for DNA and 5 for protein.
BLAT Step Size
Spacing between tiles. Default is Tile Size.
BLAT Min. Identity
Sets minimum sequence identity (in percent) between cDNA sequence and gene sequence.
BLAT Min. Score
Sets minimum score. This is the matches minus the mismatches minus some sort of gap penalty. If the the min. score is set to a high value, only long cDNA hits are taken into account.
BLAT Min. Match
Sets the number of tile matches. Usually set from 2 to 4. Default is 2 for nucleotide, 1 for protein.
BLAT Max. Gap
Sets the size of maximum gap between tiles in a clump. Usually set from 0 to 3. Default is 2. Only relevent for minMatch > 1.
BLAT One Off
If set to 1 this allows one mismatch in tile and still triggers an alignments.
| Examples | Back to top |
Drosophila melanogaster Myosin 7B gene with alternatively spliced exons
| Organism | Drosophila melanogaster |
| Genome file | |
| YAML-file for upload | |
| Query sequence | MSEFVRQHGEYVWVKPQNTTSEFAVPFGARIVRTEKTQTLVCDDRNKQFWVPAGDVLKAM HITSQEDVEDMITLGDLQEYTILRNLQNRYAKQLIYTYTGSMLVAINPYQILPIYTNREI QLYRNKSLAELPPHIFAISDNAFQRLQRLKENQCVVISGESGAGKTESTKLILQYLAAIS GKHSWIEQQIIEANPIMEAFGNAKTVRNDNSSRFGKYIEIRFTPQGAIQGARIQQYLLEK SRIVFQSRDERNYHIFYCMLAGLSTAERERLKLQEQSPSQYHYLAQGGCFTLPGRGDAKD FADIRAAMKVLSFKPEEVWSILSLLAAILHLGNLRFTATEVANLATAEIDDTPNLQRVAQ LLGIPISALNAALTQRTIFVHGEHVTTSLSKEAAIEGRDAFVKSLYDGIFVRIVRRINET INKQVDQPMNSIGVLDIFGFENFDNNSFEQLCINYANENLQQFFVGHIFKMEQDEYQNEH INWQHIEFQDNQQILDLIGMKPMNLMSLIDEESKFPKGTDQTLLEKLHVQHGNRSIYVKG KTTQTSLFGIRHYAGVVMYNPLGFLEKNRDSFSGDLRTLVQRSTNKYLVDIFPHEMPMDT AKKQPTLCVKFRNSLDMLMRTLSQAHPYFIRCIKPNEYKEPKNFDKELCVRQLRYSGMME TARIRRAGYPIRHAYRAFVERYRLLVPPVGPLEQCDCRKLARQICEVALPADSDRQYGKT KLFLRDEDDASLELQRSQLMLKSIVTIQRGIRRVLFRRYMKRYREAIITVQRYWRGRLQR RKYQVMRQGFHRLGACIAAQQLTTKFTMVRCRTIKLQALSRGYLVRKDFQKKLLERRKQN QLKKEELLKLAKMKEAEELLRLQQLKEQKEREQREQQEKRLQEEQRLKAEAAARNALAMA AVQQKRRTKPVKQEAPKAPTLQARNSLPPPPTTLIVAAPLPTRPASAVTRINTIPESPGT IDVESSKQMVDDVFRFLNDEPDAALRKLNNISSGDTIRLPKSVPNNIDTSDFSYLKYAAT YFGGGATAQHERKPLKKSLLKHEHPIDEMASKAIWLTILRFMGDLPDVVSSPTLHVFDNE NLMSDLASLLNTSDSYKPRLFVRQSQRRIPKPLASGEKEAQEFYQHWLNVPTSHLEKIHF IIGHGIIKNSLRDEILAQICKQLYLNPSRSSYSRGWLLLSLCLSCFPPSKEFEPHLRSFM KQGTAQLQATPSLQRLERTLVNGPRCQPPSLFELHAIRGRHPLRLDIHLMDGQQRRLQVD AASTAREAVNQLCQGMGLTDTFGFGLVMSLNGKLMPLGAGQEHVLDAISECEQRQLDAPW KLYIRKEMFATWYDPSMDPKATQLIYKQILNGLKCGEYRCRSEKDIAMVCALACFVEYGP GEILRLKPSEITAFVPSDLLAPGERAIENWSRLIAATYEKSSYVKEEQNDLLLEAQKRAK EDICLFAHLSWPMRHSRLFEVVRKEGPKLQSDELMLGINSAGLFLIDETEQVLASCCFSE VLKVHVESDDKLHVMTFQHVNFVLQCSSAQDANEVINYMLDNLRQRSSYGVALDPVVEGD LEDCLVLNPGDLIEFEAGVTGAQLMAGNAQDCYRGCVNGQWGQFLAGNVRVLATLTKPSE KLQDILREGRFQEPPKPTPRANYSRRRQHNISQLAESHFREPLDSDKAPLSKFSPEPLKA PLLKAVVKVPPLFQQALVMHHHILKYMGDIARSNLPVNTDLIFQPALQHPLLCDELYCQL MKQLSDNPSSESEKRGWDLLYLATGLVAPSVLVMRELIILLRMRADALADACLKRLKRSL AQGQRKKAPHLIEVEGIQQRCLHIYHKIYFPDDTVEAFEIESHTRGAELIADIAQRLELK SPVGYSIFLKTGDRVYAMPEEEFVFDFITQLIYWLRQQRTIRSISDGQYQLHFMRKLWLN NHPGEDLNGDMIFSYPQELHKYLKGYYPIDCEQASRLAILVYSADHDVSLQRLPEVLTRL IPEDLIPLQTVAEWRQQILPKVHRDHLTEDHAKILFLQELSHFACFGSTFFVVKQQNDDA LPETLLIAINSTGFHMLDPTTKEILRSYEYSQLGIWSSGKNHFHIRFGNMIGASKLLCST TQGYKMDDLLASYVKYFNEHE |
| Expert options | Gap to Close = 15 aa |
| Search for Alternatively Spliced Exons | defaults Enable search! |
| Result |

|
| Activity diagrams | Back to top |
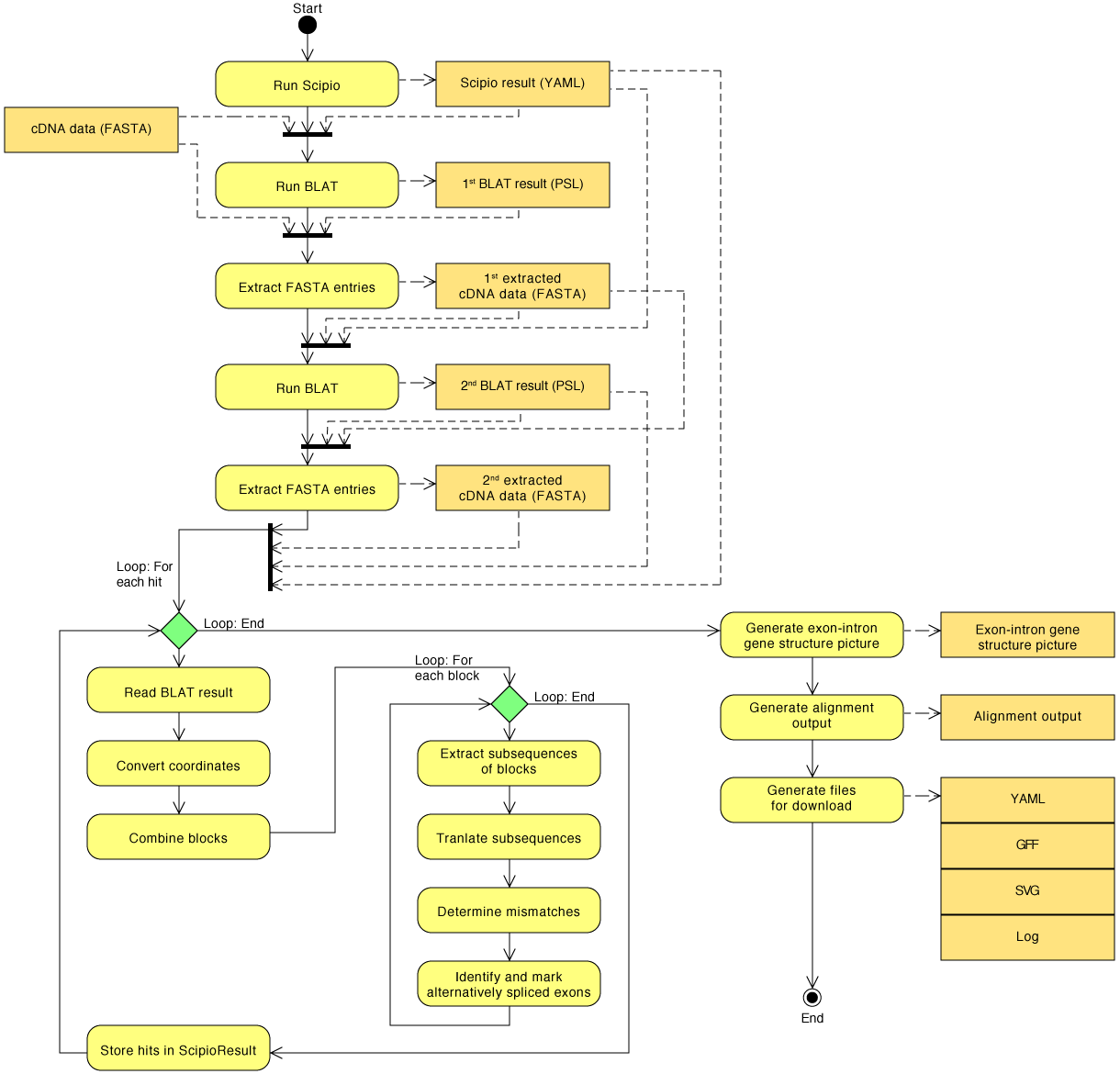
Detailed diagram of the cDNA search algorithm
|